Literature course on Evolution: All about genes, genoms and development
Posted by Louis Addo | Literature courseThis post covers chapters 14 & 15 from Futuyma and Kirkpatrick’s book on Evolution (2018). The author of this post is Jeff Marker.
The Evolution of Genes and Genomes
One of the most common sources of evolutionary adaptation is the emergence of new genes. In eukaryotes, the most common mechanism for the emergence of new genes is by gene duplication, which occurs when there is an error in DNA replication. Gene duplication can result in the creation of a new gene copy, which can then evolve to acquire new functions or to specialize in one of the functions of the original gene. The result is a gene family, which is a set of loci that originated by gene duplication and that typically have related biochemical roles. Gene families can lead to the emergence of new biochemical pathways and new structures. For example, in humans, the amylase gene family has been duplicated and evolved to produce different amylase enzymes, allowing humans to digest a wide range of starches from different plants (Perry et al., 2007).
Whole genome duplication is much rarer than gene duplication, but is a key mechanism for the evolution of the genome in many groups of organisms, especially plants. Whole genome duplication can result in the doubling of the number of chromosomes in an organism, which can then allow for the emergence of new gene functions through the duplication and divergence of gene families. For example, the multiplication of complete sets of chromosomes that have occurred in the Brassicaceae family has resulted in the emergence of many new gene functions and the evolution of novel morphological traits (Bancroft et al., 2011).
Other mechanisms of gene emergence include exon shuffling and appearance of de novo genes. Exon shuffling is a process where coding regions (exons) from different genes are rearranged and recombined, creating new genes with unique functions. De novo genes originate from non-coding regions of the genome, evolving independently of existing genes. They contribute to biological functions and evolutionary innovation through mechanisms like mutation, selection, and gene duplication.
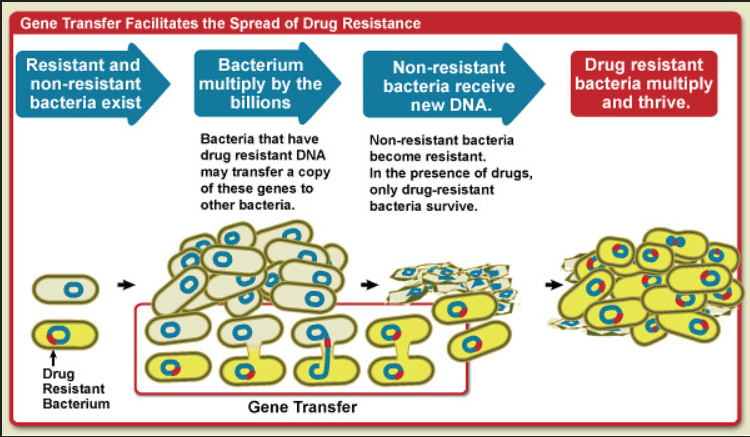
Another mechanism for the emergence of new genes is horizontal gene transfer (HGT), which involves the possession of genes from unrelated species (Figure 1). HGT is particularly common in prokaryotes, where it has enabled the rapid evolution of traits. For example, the spread of antibiotic-resistance genes among bacteria has been assisted by the HGT of plasmids carrying resistance genes (Davies and Davies, 2010).
In contrast to the previously mentioned methods of genome evolution that saw the creation of genes, chromosomal deletions can eliminate functioning genes contributing to both “good” and “bad” genomic outcomes. Natural selection can cause a deletion to increase in frequency if the deleted gene codes for a protein that increases the risk of infection. For example, a deletion in the CCR5 gene that codes for a cell surface protein used by the HIV virus to enter human cells has become fixed in some human populations.
The continuing evolution of genomes leads to a situation where some species end up with a very high number of genes. For example, the human genome consists of approximately 3 billion base pairs. However, only a small fraction of our genome is used to encode proteins and regulate gene expression. Estimates suggest that we only use about 2% of our genome to code for proteins or other gene products. The rest, termed “junk DNA,” includes regions with regulatory elements and non-functional remnants known as pseudogenes. Pseudogenes are genes that have lost their protein-coding ability through mutations. Their presence aligns with the neutral theory of molecular evolution proposed by Motoo Kimura. According to the neutral theory, the majority of genetic changes are caused by random genetic drift rather than selective pressure, resulting in non-functional sequences like pseudogenes. Understanding the significance and potential functions of these non-coding regions remains an active area of genomic research.
Evolution drives changes in gene expression patterns that can affect the survival of organisms ultimately leading to new traits and new species. Changes in gene expression occur through alterations in transcription factor binding sites, alternative splicing patterns, and epigenetic changes to DNA and histones. One example of the evolution of gene expression in insects is the wing pattern of Heliconius butterflies. Their diverse wing colors aid in reproduction and to signal to predators that they are unpalatable. Researchers found that changes in gene expression in the developmental pathway that produces the wing patterns of Heliconius butterflies have contributed to their diverse color patterns (Figure 2). The study showed that the expression of a transcription factor gene called optix, which is involved in forming the eyespot pattern on the wings, has evolved differently in different species of Heliconius butterflies (Martin et al., 2012).

Coding regions, gene expression, chromosome structure, and genome size are interconnected processes shaping an organism’s complex life. Changes in coding regions can lead to the emergence of new functional genes or alterations in protein structure. Concurrently, adjustments in gene expression patterns can drive phenotypic variation. Chromosome evolution plays a role in these processes, as rearrangements and other changes reshape the distribution of genes within a genome. Furthermore, changes in genome size can influence gene regulation, complexity, and the evolutionary trajectory of a species.
Evolution and Development
Evolutionary developmental biology (EDB) aims to integrate information from embryology, developmental genetics, and population genetics to understand how genes and changes in genes are expressed as phenotypes and changes in phenotypes. By understanding the mechanisms that produce phenotypes, we can better grasp how they evolve. Mutational changes in the genes that produce a developmental pathway may cause advantageous alterations in the phenotype, leading to the evolution of both the phenotype and its underlying genetic network.
Differences among species often result from changes in the relative developmental rates of different body parts or in the rates or durations of different life history stages, which are known as allometry and heterochrony. Some characteristics have evolved by heterotopy, where the expression occurs at a novel location on the body. For example, the inner ear bones of mammals were originally found as part of the jaw in distant common ancestors. These bones co-opted to function in the ear away from the jaw and helped mammals become more effective at hearing, aiding their survival and reproduction.
The vast diversity of multicellular eukaryotes is due to the diverse uses of a toolkit of genes and developmental pathways. Developmental pathways include signaling proteins, enhancers, transcription factors, and structural genes. Evolutionary changes in the regulatory connections among signaling pathways and transcription factors are believed to underlie much of the phenotypic diversity seen in nature. These regulatory networks coordinate the interactions among genes and other molecules to control gene expression. Regulatory networks govern biological processes and organism responses to environmental cues. One example of a gene regulatory network is that involved in the development of fruit flies. This network includes genes such as the Hox genes, which control the body segment identity, or where the different body segments are supposed to line up. The Hox genes regulate each other’s expression through a series of complex interactions, forming a network that ensures the proper patterning and differentiation of different body segments during development.
During evolution, genes and developmental pathways are often co-opted for new functions, a process that is probably responsible for the evolution of many new traits. This process results from evolutionary changes in functional connections between transcription factors and cis-regulatory elements. Modularity among body parts is achieved by patterning mechanisms whose regulation is often specific to certain structures, segments, and life history stages. Modularity helps different parts of the body develop divergent morphologies. Pleiotropic effects of genes that affect functionally interacting characteristics may evolve, resulting in the evolution of functional modules known as phenotypic integration. One example of phenotypic integration is the correlation between beak size and body size in birds. In many bird species, larger birds tend to have larger beaks, and smaller birds tend to have smaller beaks. This is an example of phenotypic integration because the size of the beak and the size of the body are functionally related traits that are influenced by overlapping sets of genes.
Genetic and developmental constraints can make some imaginable evolutionary changes unlikely. Based on changes in the expression of certain genes and developmental pathways in response to environmental signals, a single genotype may be expressed as an array of different phenotypes, known as the genotype’s norm of reaction. Reaction norms are genetically variable and can evolve by natural selection. If an environment varies, phenotypic plasticity may evolve. Conversely, selection for a constant phenotype can result in canalization. Genetic assimilation is the genetic fixation of one of the states of a phenotypically plastic character. It is not known how important genetic assimilation is in evolution, nor is it known if adaptation may occur first by a non-genetic phenotypic change that later becomes genetically fixed by natural selection.
The evolution of genes and genomes involves complex processes such as gene duplication, whole genome duplication, exon shuffling, de novo gene emergence, and horizontal gene transfer. These mechanisms contribute to the emergence of new functions and the huge amounts of genetic diversity we see in the world. Changes in gene expression patterns play a vital role in evolutionary adaptation and the development of new traits. Chromosome evolution and alterations in genome size further shape the evolutionary course of species. Understanding that these processes are firmly connected provides insights into the complexity and diversity of life forms that have evolved on our planet.
References
Bancroft, I., Morgan, C., Fraser, F., Higgins, J., Wells, R., Clissold, L., … & Trick, M. (2011). Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nature biotechnology, 29(8), 762-766.
Davies, J., & Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiology and molecular biology reviews, 74(3), 417-433.
Martin, A., Papa, R., Nadeau, N. J., Hill, R. I., Counterman, B. A., Halder, G., … & Reed, R. D. (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proceedings of the National Academy of Sciences, 109(31), 12632-12637.
Perry, G. H., Dominy, N. J., Claw, K. G., Lee, A. S., Fiegler, H., Redon, R., … & Stone, A. C. (2007). Diet and the evolution of human amylase gene copy number variation. Nature genetics, 39(10), 1256-1260.